Science Ambassador Blog: Neuroscience Gateway as a Tool for Big Data Analysis
- Details
- Published on Wednesday, 06 November 2019 14:00
One of our five Science Ambassadors, James Trevathan, Postdoctoral Fellow at the University of Wisconsin-Madison, attended OptoDBS2019 in June 2019 as well as the Neuroscience Gateways workshop on Large Scale Simulations and Data Processing in October 2019. James presented “Calcium imaging via a miniature microscope for studying deep brain stimulation in a freely moving animal model of disease.”
By James Trevathan
Thanks to the support of the SGCI Science Ambassadors, program I was able to attend and present my work at the 2019 OptoDBS conference and  later speak at the Neuroscience Gateways workshop on Large Scale Simulations and Data Processing, a satellite symposium to the Society for Neuroscience Annual Meeting. In my research, I have made use of the Neuroscience Gateway as a tool for analysis of large calcium imaging data sets collected in the striatum of unrestrained Parkinsonian mice during the application of subthalamic nucleus deep brain stimulation. While big data has transformed fields such as physics and genomics, neuroscientists and neuroengineers are increasingly collecting large scale functional and structural data and making use of high performance or high throughput computing resources for data analysis.
later speak at the Neuroscience Gateways workshop on Large Scale Simulations and Data Processing, a satellite symposium to the Society for Neuroscience Annual Meeting. In my research, I have made use of the Neuroscience Gateway as a tool for analysis of large calcium imaging data sets collected in the striatum of unrestrained Parkinsonian mice during the application of subthalamic nucleus deep brain stimulation. While big data has transformed fields such as physics and genomics, neuroscientists and neuroengineers are increasingly collecting large scale functional and structural data and making use of high performance or high throughput computing resources for data analysis.
One of the goals of the BRAIN initiative, outlined in the BRAIN 2025 report, is to “Produce a dynamic picture of the functioning brain by developing and applying improved methods for large-scale monitoring of neural activity.” [1] As large scale-monitoring techniques capable of recording from a large number of neurons within a circuit over long periods of time become available to individual neuroscience laboratories, the difficulty becomes understanding and exploiting the full potential of these complex high-dimensional data. My experience in this area began during my PhD within the laboratory of Luis Lujan at the Mayo Clinic. Dr. Lujan’s laboratory uses a combination of neurophysiology techniques and modeling approaches to understand the effects of deep brain stimulation. Although deep brain stimulation is an approved therapy for the treatment of Parkinson’s’ Disease, essential tremor, dystonia, and obsessive-compulsive disorder the mechanism by which deep brain stimulation achieves therapeutic effect remain unclear. Looking for a new approach to understand the circuit level effects of deep brain stimulation therapy I began using calcium imaging, a technique that takes advantage of genetically encoded fluorescent calcium indicators to measure changes in intracellular calcium as a surrogate for neural activity. We performed these recordings using a head-mounted miniature fluorescent microscope, an increasingly popular technique for simultaneously recording from hundreds of neurons in awake behaving small animal models. Additionally, this method allowed us to record, through an implanted microendoscope, from deep brain regions of the basal ganglia circuit affected by DBS. During these experiments, I moved from working with data sets that were on the order of gigabytes to collecting terabytes of data from a cohort of animals. In total, the first paper we submitted[2] using the technique included analysis performed on around 15 terabytes of raw data. The computational resources available within our laboratory were not well optimized for working with this amount of data, so I began to seek out options for either expanding our computational infrastructure, which was potentially expensive, or utilizing high performance computing resources.
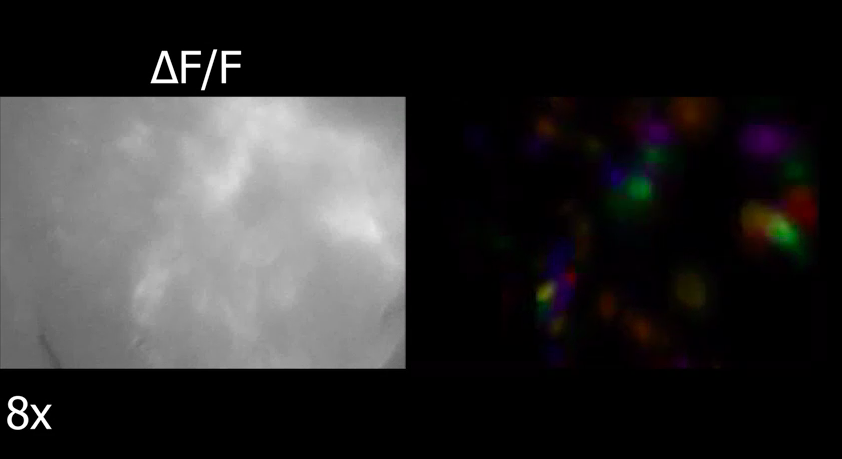
Screenshot of calcium imaging performed in one of Trevathan's experiments.
Looking for solutions to our data analysis bottleneck, I found and started working with the Neuroscience Gateway. Although the Neuroscience Gateway was originally designed as a resource for the computational neuroscience community and focused on tools for simulation and modeling, it is also well suited for data processing. In my case, I primarily used the gateway for running Constrained Non-negative Matrix Factorization for Microendoscope data (CNMF-E)[3], a published algorithm for extracting the location of neuronal cell bodies as well as calcium traces, indicative of neuronal activity, from calcium imaging videos. CNMF-E and other algorithms for identification of neuronal activity from calcium imaging data sets generally act as a way to extract relevant features from high dimensional imaging data that can be further processed to draw conclusions across data sets. Prior to leveraging the Neuroscience Gateway, performing CNMF-E on a 30-minute recording of neuronal activity obtained within our laboratory took close to 24 hours on a high-end workstation. Since CNMF-E, published in Matlab, could be run on the Neuroscience Gateway using Matlab and process a single data set per compute node, the gateway allowed for parallelizing our data processing across the dozens of data sets that we had collected. This moved the bottleneck in our research from running CNMF-E to uploading data to the Neuroscience Gateway. This allowed me to focus my time and the computational resources within our laboratory on drawing conclusions about the effects of deep brain stimulation from the neuronal activity traces identified by CNMF-E.
While at the Neuroscience Gateway workshop just before the Society for Neuroscience annual meeting last month, I had the opportunity to learn about upcoming features and directions for the gateway. The Neuroscience Gateway, because it was originally designed primarily for computational neuroscience applications requiring high performance computing capabilities for simulation and modeling, allows for uploading and working with large data sets but is not well optimized for this application. With the increase of big data applications making use of the Neuroscience Gateway for processing of a variety of imaging or electrophysiology data types, the gateway developers are working on improving capabilities for uploading and working with large data sets. I am looking forward to these improvements and will be continuing to use the Neuroscience Gateway for future projects as I move into a new postdoctoral fellowship position at the University of Wisconsin-Madison.
As other neuroscience and neuroengineering groups begin collecting large data sets that outpace their ability to perform analysis using the computational resources available within their laboratory, I would encourage them to look at the resources available through the Neuroscience Gateway. The built-in tools, libraries, and pipelines are an easy way to get started using the gateway. However, making use of custom Matlab (Mathworks; Natick, MA) or Python code on the high throughput computing resources available through the gateway provides an option for performing custom data analysis or running published code on large data sets. In this way, using the Neuroscience Gateway can significantly accelerate the data analysis process.